griddata¶
-
mipylib.numeric.minum.griddata(points, values, xi=None, **kwargs)¶ Interpolate scattered data to grid data.
- Parameters
points – (list) The list contains x and y coordinate arrays of the scattered data.
values – (array_like) The scattered data array.
xi – (list) The list contains x and y coordinate arrays of the grid data. Default is
None, the grid x and y coordinate size were both 500.method – (string) The interpolation method. [idw | cressman | neareast | inside | inside_min | inside_max | inside_count | surface]
fill_value – (float) Fill value, Default is
nan.pointnum – (int) Only used for ‘idw’ method. The number of the points to be used for each grid value interpolation.
radius – (float) Used for ‘idw’, ‘cressman’ and ‘neareast’ methods. The searching raduis. Default is
Nonein ‘idw’ method, means no raduis was used. Default is[10, 7, 4, 2, 1]in cressman method.convexhull – (boolean) If the convexhull will be used to mask result grid data. Default is
False.
- Returns
(array) Interpolated grid data (2-D array)
Examples
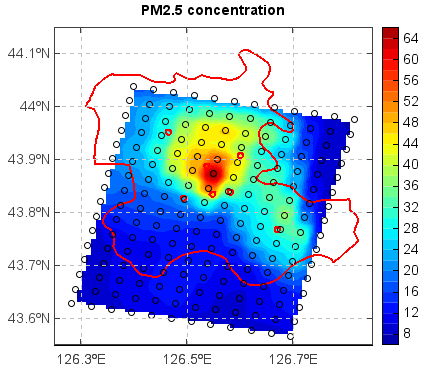
f = addfile('D:/temp/nc/out.20140421_20140421_JL3KMmeic.nc') data = f['PM25'] data = data[15,1,:,:] lon = f['lon'][:,:] lat = f['lat'][:,:] #Interpolate data to grid lon1 = linspace(lon.min(), lon.max(), lon.dimlen(1)*5) lat1 = linspace(lat.min(), lat.max(), lat.dimlen(0)*5) data1 = griddata((lon, lat), data, xi=(lon1, lat1), method='idw', convexhull=True)[0] lon_g,lat_g = meshgrid(lon1, lat1) #Plot axesm() mlayer = shaperead('D:/temp/map/jilin.shp') geoshow(mlayer, edgecolor='r', size=2) layer = contourfm(lon1, lat1, data1, 20) scatterm(lon, lat, data, 20, fill=False) colorbar(layer) xlim(126.25,126.85) ylim(43.55,44.15) grid(True) title('PM2.5 concentration')