Write netCDF data file¶
Several steps are needed to create a netCDF data file and write data in it:
Create a writable data file object using
addfile()function. The first argument is
file name and the second one is 'c' which means creating data file.
Add dimensions using
adddim()function of the data file object. The two arguments
are dimension name and length.
Add global attributes using
addgroupattr()function of the data file object. The
two arguments are attribute name and value.
Add variables using
addvar()function of the data file object. The three arguments
are variable name, data type and dimensions.
Create netCDF file using
create()function of the data file object.Write data array to the netCDF file using
write()function of the data file object.Close netCDF data file by
close()function of the data file object.
The below example shows reading 4 netCDF files and joint them into a new netCDF data file.
datadir = 'D:/Temp/nc'
outfn = os.path.join(datadir, 'join_file.nc')
#New netCDF file
ncfile = addfile(outfn, 'c')
#Add dimensions
stn = 26564
recdim = ncfile.adddim('recNum', stn)
stdim = ncfile.adddim('station', stn)
iddim = ncfile.adddim('id_len', 11)
tdim = ncfile.adddim('time', 4)
#Add global attributes
ncfile.addgroupattr('Conventions', 'Unidata Observation Dataset v1.0')
ncfile.addgroupattr('cdm_datatype', 'Station')
ncfile.addgroupattr('geospatial_lat_max', '90.0')
ncfile.addgroupattr('geospatial_lat_min', '-90.0')
ncfile.addgroupattr('geospatial_lon_max', '180.0')
ncfile.addgroupattr('geospatial_lon_min', '-180.0')
ncfile.addgroupattr('stationDimension', 'station')
ncfile.addgroupattr('missing_value', -8.9999998E15)
ncfile.addgroupattr('stream_order_output', 1)
#Add variables
variables = []
var = ncfile.addvar('latitude', 'float', [stdim]) #Latitude
var.addattr('long_name', 'station latitude')
var.addattr('units', 'degrees_north')
variables.append(var)
var = ncfile.addvar('longitude', 'float', [stdim]) #Longitude
var.addattr('long_name', 'station longitude')
var.addattr('units', 'degrees_east')
variables.append(var)
var = ncfile.addvar('altitude', 'float', [stdim]) #Altitude
var.addattr('long_name', 'station altitude')
var.addattr('units', 'meters')
variables.append(var)
var = ncfile.addvar('streamflow', 'float', [tdim, stdim]) #Stream flow - Add time dimension
var.addattr('long_name', 'River Flow')
var.addattr('units', 'meter^3 / sec')
variables.append(var)
tvar = ncfile.addvar('time', 'int', [tdim])
tvar.addattr('long_name', 'time')
tvar.addattr('units', 'hours since 1900-01-01 00:00:0.0')
#Creat netCDF file
ncfile.create()
#Write data
stime = datetime.datetime(2015,8,2,0)
etime = datetime.datetime(2015,8,2,3)
st = datetime.datetime(1900,1,1)
fi = 0
while stime <= etime:
print stime
fn = os.path.join(datadir, stime.strftime('%Y%m%d%H') + '00.CHRTOUT_DOMAIN2')
if os.path.exists(fn):
print '\t' + fn
f = addfile(fn)
hours = (stime - st).total_seconds() // 3600
origin = [fi]
ncfile.write(tvar, array([hours]), origin=origin)
if fi == 0:
lat = f['latitude'][:]
ncfile.write(variables[0], lat)
lon = f['longitude'][:]
ncfile.write(variables[1], lon)
alt = f['altitude'][:]
ncfile.write(variables[2], alt)
flow = f['streamflow'][:]
origin = [fi, 0]
shape = [1, stn]
flow = flow.array.reshape(shape)
ncfile.write(variables[3], flow, origin=origin)
fi += 1
stime = stime + datetime.timedelta(hours=1)
#close netCDF file
ncfile.flush()
ncfile.close()
print 'Finished!'
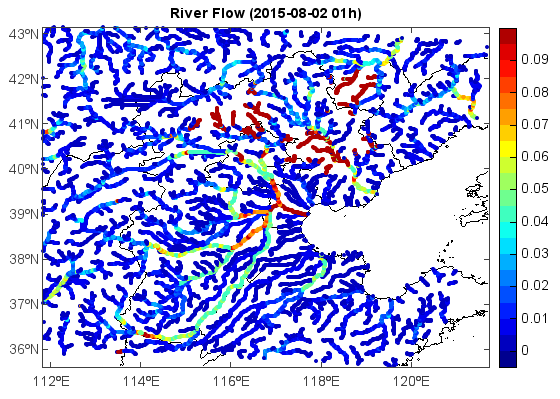
Read and plot joined netCDF data file:
f = addfile('D:/Temp/nc/join_file.nc')
lon = f['longitude'][:]
lat = f['latitude'][:]
var = f['streamflow']
flow = var[1,:]
axesm()
geoshow('cn_province')
levs = arange(0, 0.1, 0.005)
layer = scatter(lon, lat, flow, levs, edge=False)
colorbar(layer)
t = f.gettime(1)
title('River Flow (' + t.strftime('%Y-%m-%d %Hh)'))